From Slides to Screens: Virtual Staining in Action
Take a closer look at the behind-the-envelope work in virtual staining
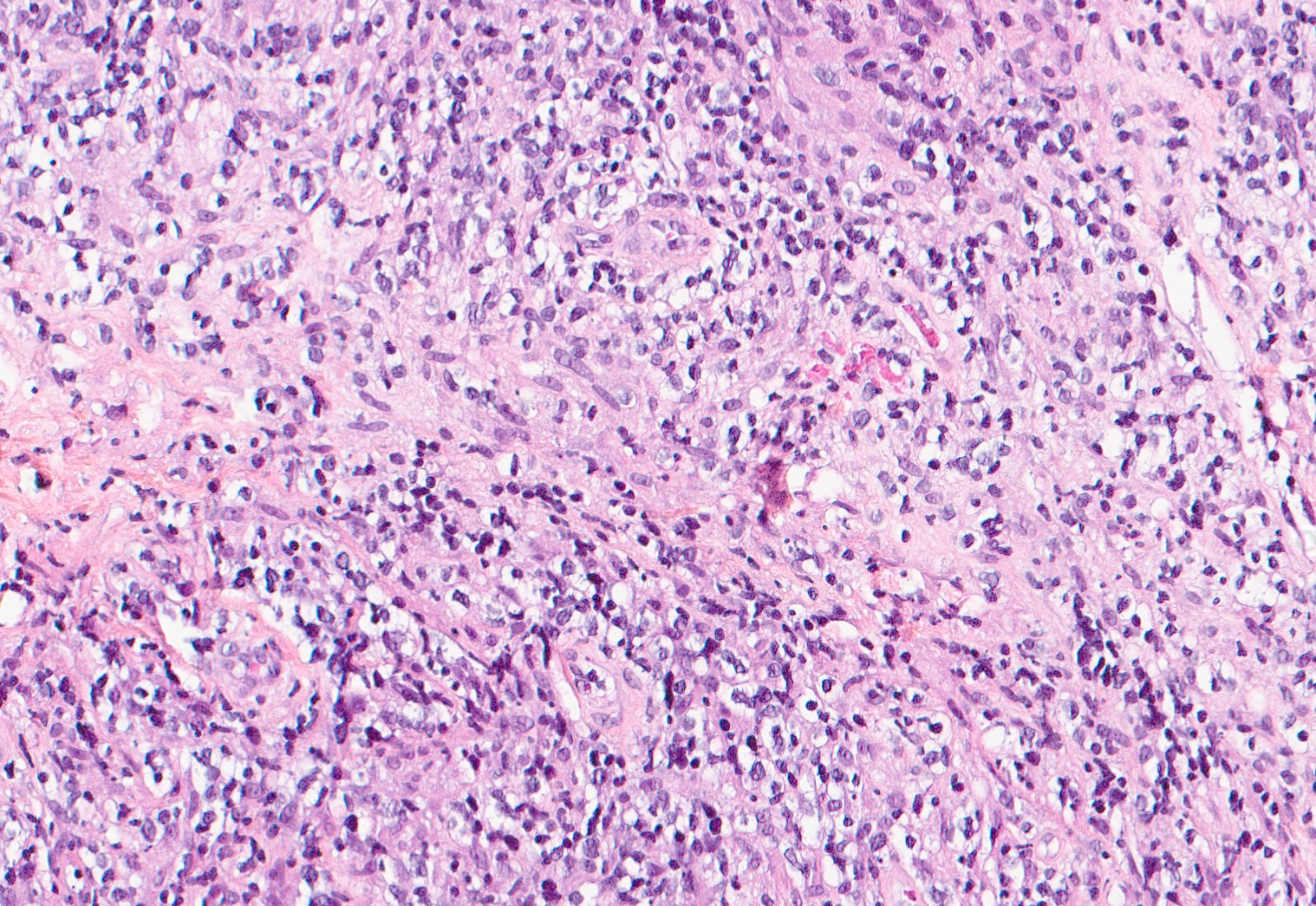
With a backbone comprised of deep learning, virtual staining algorithms are trained on thousands of paired tissue images. This is how by just reading the unstained biopsy slide scans, the virtual staining software generates predicted H&E and IHC slides on your screen.
By bridging cutting-edge R&D with real-world deployment, we help laboratories save time, cut chemical agents usage and create values.
Label-Free Staining
The first class of virtual staining is called label-free staining. It turns autofluorescence images of unstained samples to H&E stain.
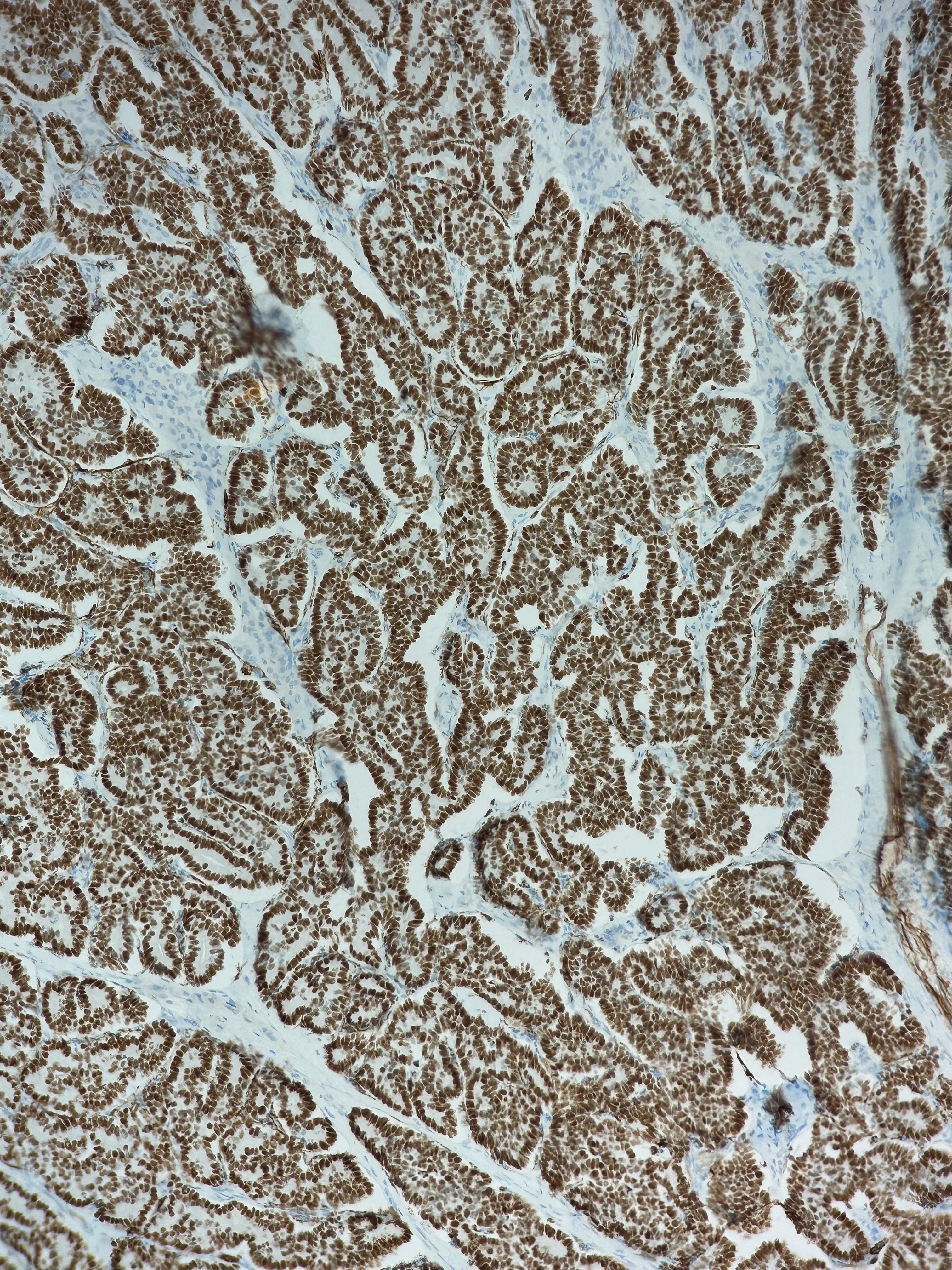
Stain-to-Stain Translation
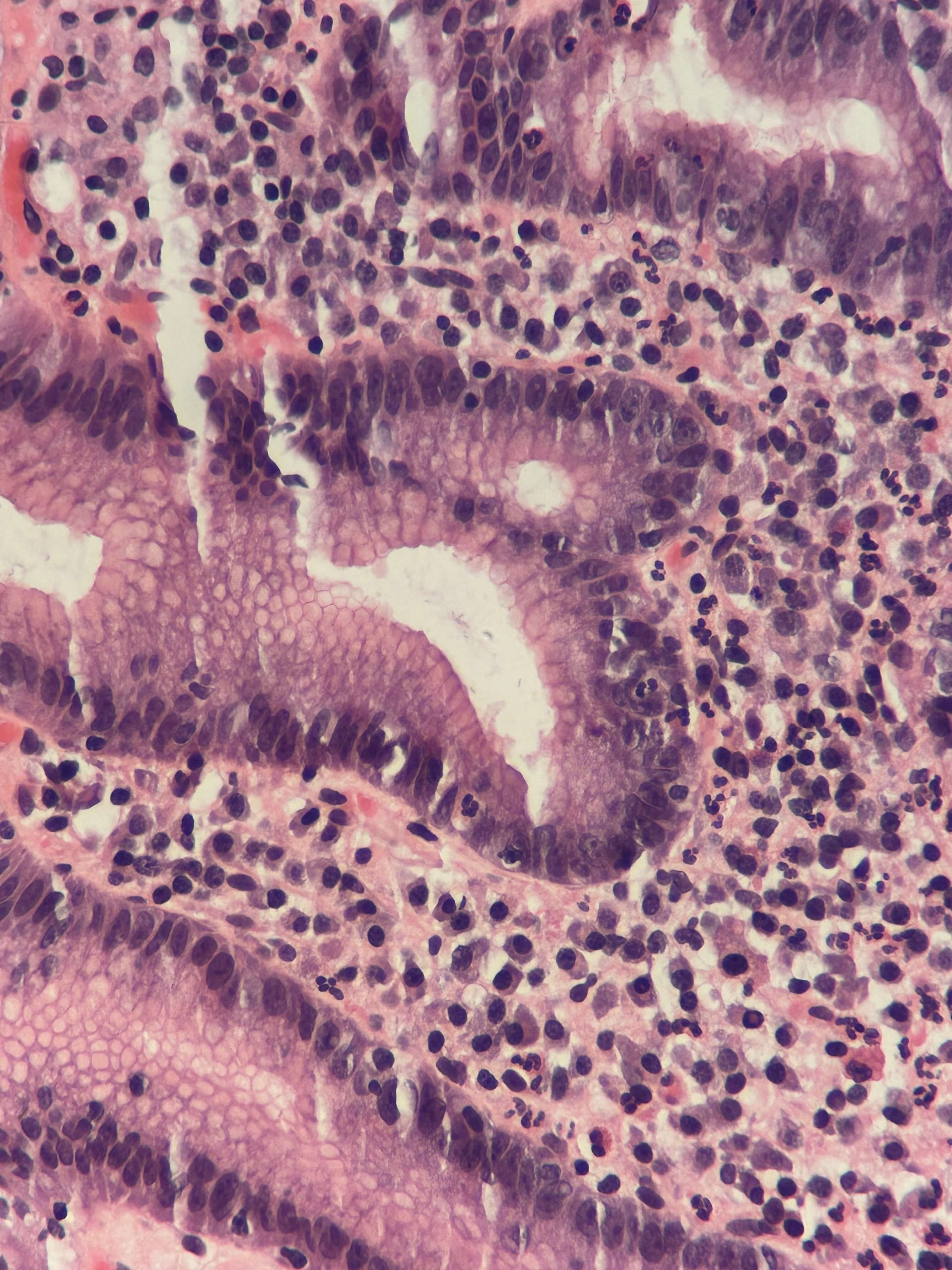
As its name suggests, the second class of virtual staining converts one type of stain to another. The most common conversion is from H&E to IHC stain.
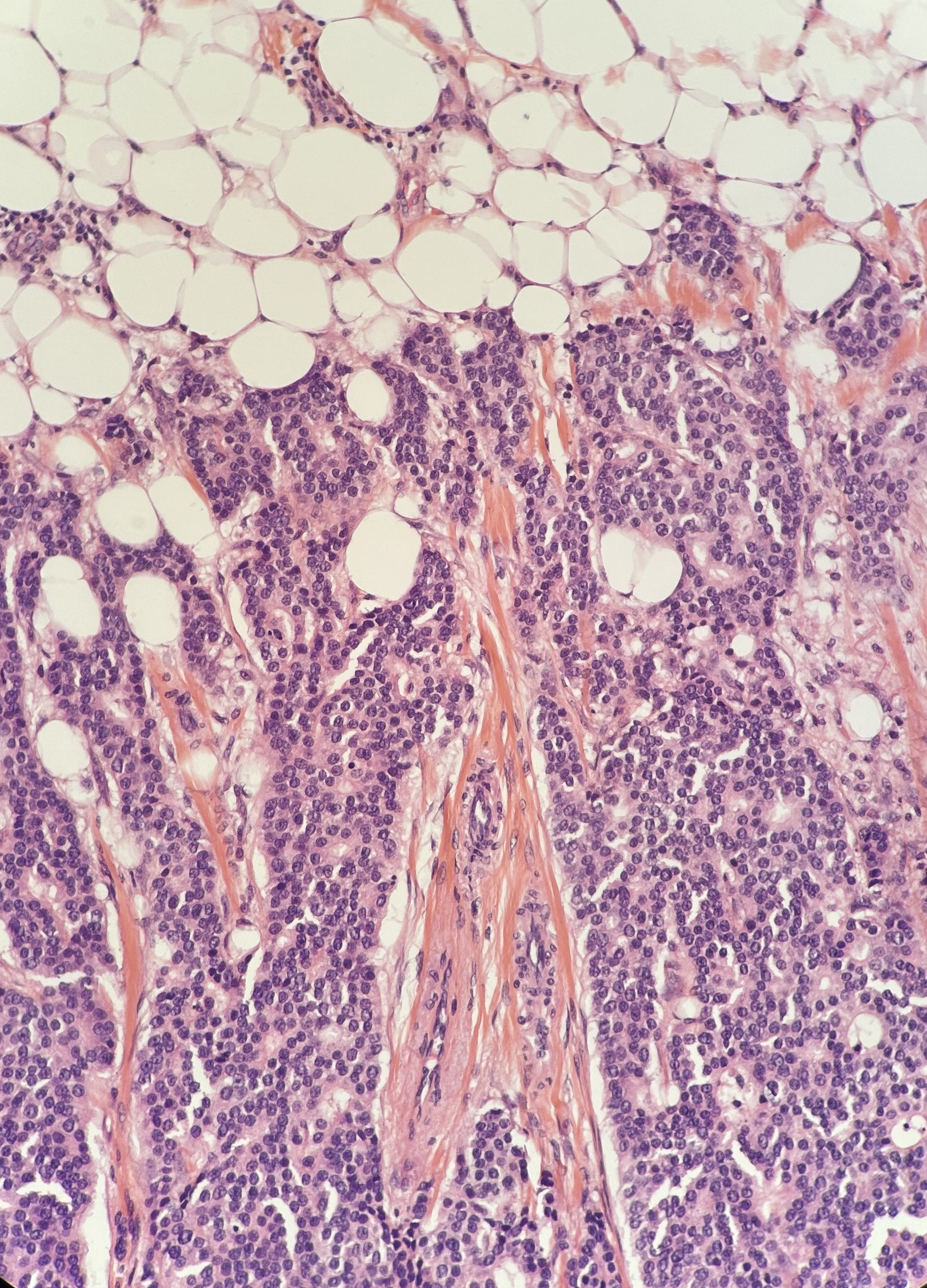
In-silico Multiplex Staining
This third class of virtual staining predicts the expression of multiple protein biomarkers from H&E images. It enables computational cell phenotyping and microenvironment analysis.
We deliver trusted AI technologies for smarter, faster pathology.
Explore our scientific paper digest here!
Validated precision (SSIM)
Using a dataset of 523 samples, Zhang et al. demonstrated that label-free staining can reliably generate high-quality H&E stain predictions with strong structural similarity as physical stains.
Pathologists Review
In a double-blind study, pathologists compared real and virtually stained slides from 250 samples, confirming that the AI-generated stains produced similar diagnoses with high structural and perceptual quality (De Haan et al., 2021).
Reference
De Haan, K. et al. Deep learning-based transformation of H&E stained tissues into special stains. Nat Commun 12, 4884 (2021).
Zhang, Y. et al. Digital synthesis of histological stains using micro-structured and multiplexed virtual staining of label-free tissue. Light Sci Appl 9, 78 (2020).
High quality output with precision
Using breast cancer biopsy samples, Klöckner et al. (2025) validated a deep learning pipeline that translates H&E into virtual IHC stains (ER, PR, HER2, Ki-67).
Across multiple test sets, the model showed strong structural and perceptual similarity to real IHC, with PSPStain achieving the best performance.
Reference
Klöckner, P. et al. H&E to IHC virtual staining methods in breast cancer: an overview and benchmarking. npj Digit. Med. 8, 384 (2025).
Validated Clinical Relevance Across Sites
Using 4 independent datasets, the algorithm showed robust prediction of 50 protein biomarkers from H&E slides.
Across datasets, the algorithm significantly outperforming baseline H&E- and morphology-based models.
Cell phenotyping analysis further showed accurate classification of 7 cell types, with performance comparable to ground-truth data.
Reference
Wu, E. et al. ROSIE : AI generation of multiplex immunofluorescence staining from histopathology images. Preprint at https://doi.org/10.1101/2024.11.10.622859 (2024).
Precision detection of biomarkers on any slide...
Why it matters ?
Accurate biomarker detection is the foundation of precision medicine. By identifying the right molecular signals, pathologists can deliver more personalised, targeted cancer care.
At RainPath, our AI-powered software suite is designed to support your lab, strengthen diagnostic confidence, and prepare you for the era of precision medicine.




See our used case of biomarker identification: Ki-67
Why is the new trend of AI and digital pathology reliable?
Explore our news digest of AI healthcare cases in real life!
AI models for virtual staining are built on big training data sets of 100+ and 1000+ of paired biopsy slide images.
Models are validated with internal and external cohorts of data as shown in the previous slides.
Validated by certified pathologists on multiple organ sites including salivary gland, thyroid, kidney, liver and lung (Rivenson et al., 2019).
OSAIRIS:
Addenbrooke’s Hospital, UK
Dr Raj Jena and team used AI to automatically segment organs, reducing radiotherapy planning time by 2.5x from hours to minutes.
iSeg:
Northwestern University, USA
Dr Abazeed and Dr Teo deployed a 3D deep learning model to segment lung tumors in moving lungs for radiation planning, improving tumour detection accuracy.
Sarkar, S., Teo, P. T. & Abazeed, M. E. Deep learning for automated, motion-resolved tumor segmentation in radiotherapy. npj Precis. Onc. 9, 173 (2025).
NHS, UK:
The NHS (UK) deployed an AI tool that analyses brain scans across 107 stroke centres in England.
Outcome: Increasing the full recovery rates from 16% to 48%.
NewYork–Presbyterian Hospital, USA:
Dr Poterucha and his team developed a deep learning model (EchoNext) trained on over 1 million ECG + imaging records.
Outcome: Through controlled external validation studies, they showed that EchoNext can identify structural heart disease with a higher accuracy than cardiologists.
Poterucha, T. J. et al. Detecting structural heart disease from electrocardiograms using AI. Nature 644, 221–230 (2025).

.svg.png)